Advances in deep learning of representation have resulted in powerful generative approaches on modeling continuous data like time series and images, but it is still challenging to correctly deal with discrete structured data, such as chemical molecules and computer programs. To tackle these challenges, there has been many improvements in formalization of structure generation that help deep models better model complex structured under constraints of limited data. Improving previous works that formalize data as arbitrary string (CVAE) and generated string from context free grammar (GVAE), we propose a new formalization framework that allows both syntactical and semantic constraints of structured data to be used for guiding the data modeling. Our approach facilities a generative model that models data in a much more powerful way with both syntax and semantics constraints.
In our recent paper Syntax-Directed Variational Autoencoder for Structured Data accepted at ICLR 2018, we detailed our new formalization, modeling and experiments. Simply put, our proposed formalization, or SD-VAE, reshapes the output space of generative model more tightly to the meaningful data space, as show in Figure 1, by systematically converting the offline semantic check into online guidance for generating.
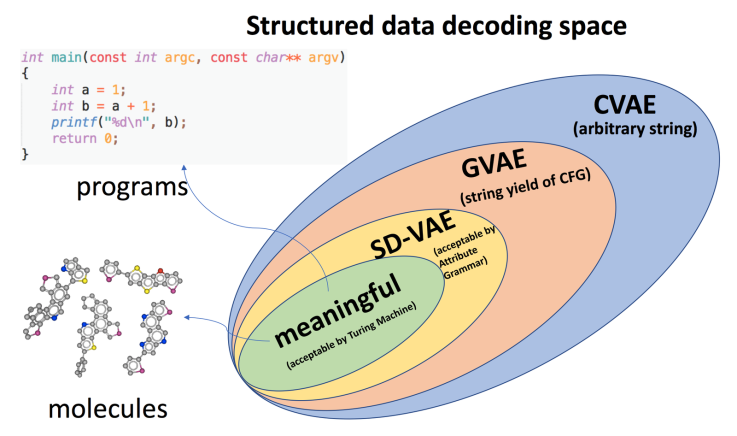
Even with the increased capacity, our approach has the same computational cost as previous models for efficient learning and inference. Finally it results in significant and consistent improvements across various tasks on chemical molecules and computer programs, showing its empirical value of solving real world problem.
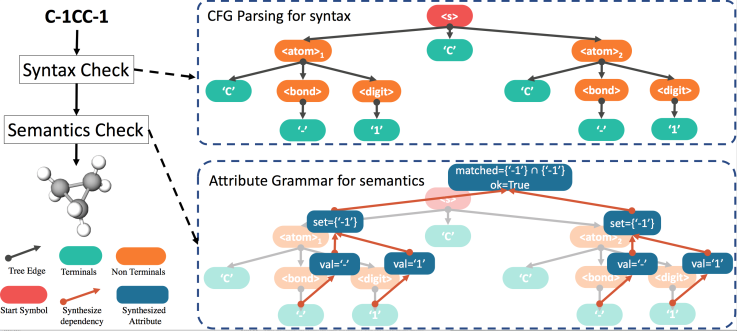
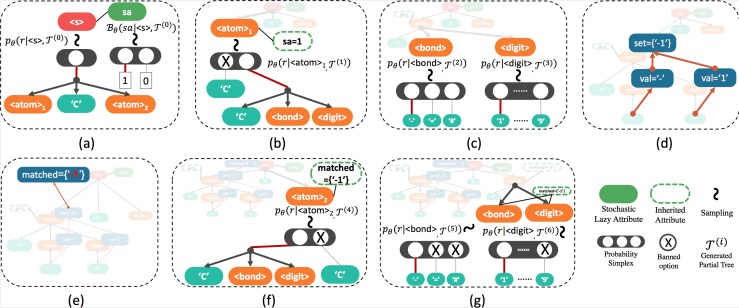
Our method converts the offline Attribute Grammar, an shown in Figure 2, to on-the-fly generative process. In Attribute Grammar, there are inherited and synthesized attribute that are computed offline for ensuring semantic correctness. To enable the on-the-fly computation during tree generation we introduce the stochastic lazy attributes that transforms the corresponding synthesized attribute into inherited constraints in generative procedure; and a lazy linking mechanism sets the actual value of the attribute, once all the other dependent attributes are ready, as illustrated in Figure 3.
Our formalization works with any generative model, but in practice we choose to have it work with Variational Autoencoder, or VAE, which is an explicit generative model that converts between discrete data space and continuous latent space, for fair comparison with previous models. Empirical results show that our formalization boosts performance in reconstruction accuracy and valid prior accuracy, both important quality metric for VAE, as shown in Figure 4.

Following GVAE, we use Bayesian Optimization to find new molecules with desired drug-related property. Our method offers molecules with much better drug-likeliness than previous models, as shown in Figure 5, and similarly computer programs that are close to a particular ground truth, as in Figure 6.


the right are top 3 closest programs found by each method along with the distance to ground truth. Lower distance is better, and our method finds symbolic program, when used as a function, closest to the ground truth.
A few other experiments also suggest that our approaches models the data in a way that similar data are placed near each other (Figure 7) and models diversely, which are desired priorities of good generative model.

While our work is the first to tackle challenges of address both syntax and semantics with prior knowledge, and shows strong imperial improvements, there are still more problems to tackle. For example, we would like to explore the refinement of formalization on a more theoretical ground, and investigate the application of such formalization on a more diverse set of data modality.
[Post by: Hanjun Dai*, Yingtao Tian*, Bo Dai, Steven Skiena, Le Song]
